GRETEL
Is an open source framework for Evaluating Graph Counterfactual Explanation Methods. It is implemented using the Object Oriented paradigm and the Factory Method design pattern. Our main goal is to create a generic platform that allows the researchers to speed up the process of developing and testing new Graph Counterfactual Explanation Methods. GRETEL provides all the necessary building blocks to create bespoke explanation pipelines.
Latest version (v2.0) of the GRETEL Framework is available here.
GRETEL Framework (v1.0) is available here.
Why Counterfactual Explanations on Graphs?
Machine Learning (ML) systems are a building part of the modern tools that impact our daily lives in several application domains. Graph Neural Networks (GNN), in particular, have demonstrated outstanding performance in domains like traffic modeling, fraud detection, large-scale recommender systems, and drug design. However, due to their black-box nature, those systems are hardly adopted in application domains where understanding the decision process is of paramount importance (e.g., health, finance). Explanation methods were developed to explain how the ML model has taken a specific decision for a given case/instance. Graph Counterfactual Explanations (GCE) is one of the explanation techniques adopted in the Graph Learning domain. GCEs provide explanations of the kind “What changes need to be done in the graphs to change the prediction of the GNN.” Counterfactuals provide recourse to users, allowing them to take actions to change the outcomes of the decision systems while allowing the developers to identify bias and errors in the models. The following figure shows how conterfactual explanations can be used in drug discovery be identifying molecular structures associated to undesired effects and changing them transforming cephallexin into amoxicillin.
Figure 1: Example on the drug discovery task.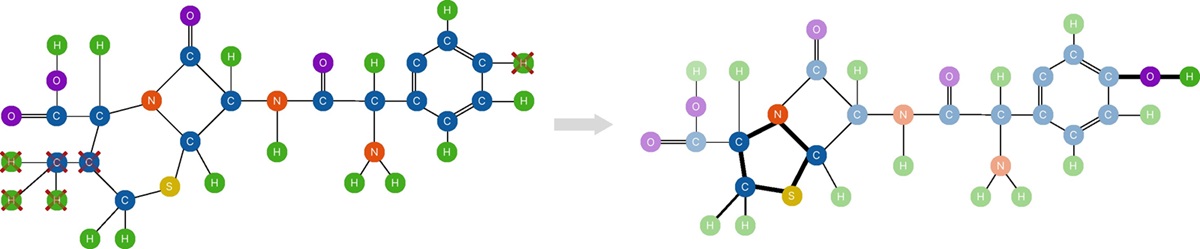
Resources included by the Framework:
GRETEL offers many out-of-the-box components that facilitate the use of the framework for creating custom-made explanation pipelines without the need to implement features beyond the user’s interest.
Datasets:
-
Tree-Cycles [3]: Synthetic data set where each instance is a graph. The instance can be either a tree or a tree with several cycle patterns connected to the main graph by one edge
-
Tree-Infinity: It follows the approach of the Tree-Cycles, but instead of cycles, there is an infinity shape.
-
ASD [4]: Autism Spectrum Disorder (ASD) taken from the Autism Brain Imagine Data Exchange (ABIDE).
-
ADHD [4]: Attention Deficit Hyperactivity Disorder (ADHD), is taken from the USC Multimodal Connectivity Database (USCD).
-
BBBP [5]: Blood-Brain Barrier Permeation is a molecular dataset. Predicting if a molecule can permeate the blood-brain barrier.
-
HIV [5]: It is a molecular dataset that classifies compounds based on their ability to inhibit HIV.
Oracles:
-
KNN
-
SVM
-
GCN
-
ASD Custom Oracle [4] (Rules specific for the ASD dataset)
-
Tree-Cycles Custom Oracle (Guarantees 100% accuracy on Tree-Cycles dataset)
Explainers:
-
DCE Search: Distribution Compliant Explanation Search, mainly used as a baseline, does not make any assumption about the underlying dataset and searches for a counterfactual instance in it.
-
Oblivious Bidirectional Search (OBS) [4]: It is an heuristic explanation method that uses a 2-stage approach.
-
Data-Driven Bidirectional Search (DDBS) [4]: It follows the same logic as OBS. The main difference is that this method uses the probability (computed on the original dataset) of each edge to appear in a graph of a certain class to drive the counterfactual search process.
-
MACCS [5]: Model Agnostic Counterfactual Compounds with STONED (MACCS) is specifically designed to work with molecules.
-
MEG [6]: Molecular Explanation Generator is an RL-based explainer for molecular graphs.
-
CFF [7] Is a learning-based method that uses Counterfactual and Factual Reasoning in the perturbation mask generation process.
-
CLEAR [8] is a learning based explanation method that provides Generative Counterfactual Explanations on Graphs.
-
G-CounteRGAN [9] is a porting of a GAN-based explanation method for images
References:
-
Prado-Romero, M.A. and Stilo, G., 2022, October. Gretel: Graph counterfactual explanation evaluation framework. In Proceedings of the 31st ACM International Conference on Information & Knowledge Management (pp. 4389-4393).
-
Prado-Romero, M.A., Prenkaj, B. and Stilo, G., 2023, February. Developing and Evaluating Graph Counterfactual Explanation with GRETEL. In Proceedings of the Sixteenth ACM International Conference on Web Search and Data Mining (pp. 1180-1183).
-
Zhitao Ying, Dylan Bourgeois, Jiaxuan You, Marinka Zitnik, and Jure Leskovec. 2019. Gnnexplainer: Generating explanations for graph neural networks. Ad- vances in neural information processing systems 32 (2019)
-
Carlo Abrate and Francesco Bonchi. 2021. Counterfactual Graphs for Explainable Classification of Brain Networks. In Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining. 2495–2504
-
Geemi P Wellawatte, Aditi Seshadri, and Andrew D White. 2022. Model agnostic generation of counterfactual explanations for molecules. Chemical science 13, 13 (2022), 3697–370
-
Numeroso, D. and Bacciu, D., 2021, July. Meg: Generating molecular counterfactual explanations for deep graph networks. In 2021 International Joint Conference on Neural Networks (IJCNN) (pp. 1-8). IEEE.
-
Tan, J., Geng, S., Fu, Z., Ge, Y., Xu, S., Li, Y. and Zhang, Y., 2022, April. Learning and evaluating graph neural network explanations based on counterfactual and factual reasoning. In Proceedings of the ACM Web Conference 2022 (pp. 1018-1027).
-
Ma, J., Guo, R., Mishra, S., Zhang, A. and Li, J., 2022. Clear: Generative counterfactual explanations on graphs. Advances in Neural Information Processing Systems, 35, pp.25895-25907.
-
Nemirovsky, D., Thiebaut, N., Xu, Y. and Gupta, A., 2022, August. CounteRGAN: Generating counterfactuals for real-time recourse and interpretability using residual GANs. In Uncertainty in Artificial Intelligence (pp. 1488-1497). PMLR.



